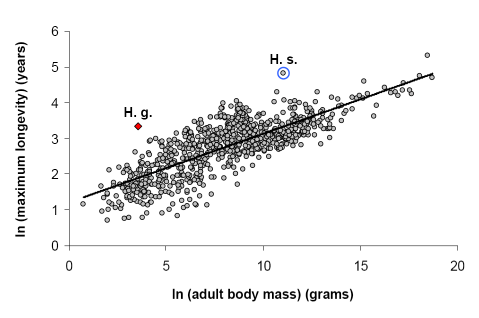
What is the naked mole-rat?

The naked mole-rat (Heterocephalus glaber) is a long-lived subterranean rodent from the Horn of Africa. The general appearance is that of a small (8 to 10cm long) wrinkled pink animal with overlarge front teeth. The naked mole-rat lives in an underground burrow system that has led to a number of adaptions such as the general inability to regulate body temperature (underground temperature is relatively stable), low metabolic and respiratory rates and lack of reaction to pain in the animals' skin.
They live in a colony with a complex social structure, ranging in size from 75 to 200 individuals. There is a single breeding queen and one to three breeding males. The other individuals in the colony are temporary sterile workers that collect food and defend the colony. Once the queen dies one of the sterile females will take over the position, losing its sterility.